Submitted by yuzhounh on Sun, 07/20/2014 - 16:00
Dear all,
I have downloaded the test data from http://www.restfmri.net/forum/DemoData and load the configuration data DPARSF_Preprocess_ALFF_FC.mat by DPARSF v2.1 like below
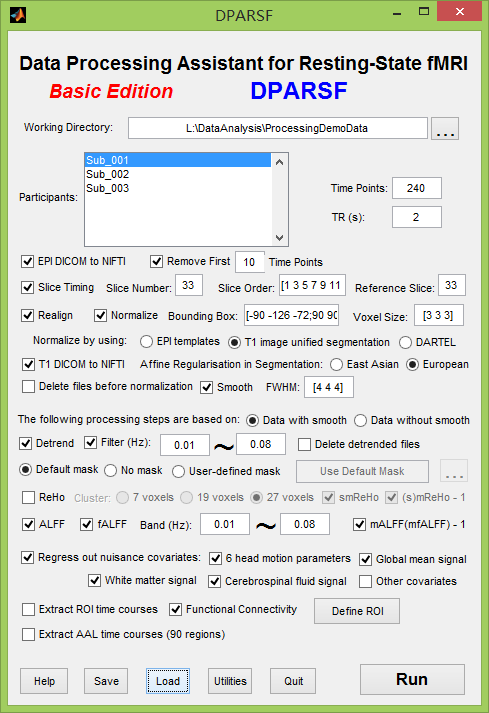
An error occurs in the step of "Regress out nuisance covariates" like below. The data in the folder of FunImgNormalizedSmoothedDetrendedFilteredCovs must be wrong. Could you tell me what's the problem?
Error using horzcat
Dimensions of matrices being concatenated are not consistent.
Error in DPARSF_run (line 1209)
Covariables=[Covariables,theROITimeCourses];
Error in DPARSF>pushbuttonRun_Callback (line 976)
[Error]=DPARSF_run(handles.Cfg);
Error in gui_mainfcn (line 96)
feval(varargin{:});
Error in DPARSF (line 43)
gui_mainfcn(gui_State, varargin{:});
Error while evaluating uicontrol Callback
Thank you very much for your help!
Best regards,
Jing.

Submitted by gaolei6096 on Wed, 07/23/2014 - 15:10 Permalink
Re: Problem of DPARSF v2.1
Hi, you can reference the following solution:
http://www.restfmri.net/forum/node/1353
"Fri, 02/01/2013 - 02:49 — YAN Chao-Gan
Re: Error using horzcat - Seed-based Functional Connectivity
Wed, 01/30/2013 - 19:02 — zangzx
Re: Error using horzcat - Seed-based Functional Connectivity
Hi,
Covariables=[Covariables,theROITimeCourses];
You need to make sure that your covariates and ROI series are equal length. If the ROI time course contains 100 time points, then your covariates must have 100 time points too.
Submitted by yuzhounh on Thu, 07/24/2014 - 00:49 Permalink
Re: Problem of DPARSF v2.1
Thank you for your help.
But I do not quite understand how this problem occurs since the data and configuration I have tried are provided by the authors of DPARSF. The problem surely relates to the command of [Covariables, theROITimeCourses], but how to solve this problem since DPARSF is run under GUI?
There are totally 240 time points and 10 of them are removed. In the rp*.txt, the data size is 230*6. There is no problem till this step.
The files in the folder of FunImgNormalizedSmoothedDetrendedFilteredCovs are wrong. How could I correct them?
Thanks!
Jing.
Submitted by yuzhounh on Thu, 07/24/2014 - 00:53 Permalink
Re: Problem of DPARSF v2.1
Also, "Yeah, please make sure at which step you removed some first time points." How could I determine or change this? The order of steps seems to be fixed on the GUI. Jing.
Submitted by gaolei6096 on Sat, 07/26/2014 - 01:29 Permalink
Re: Problem of DPARSF v2.1
What you would usually do is to remove the generated folders and files, rerun the DPARSF parameters configuration, and then it should help.