troubles while processing DPARSF basic edition, never happened before
Dear REST experts:
Recently I run the DPARSF basic edition and I faced the following wrong messages in my matlab command window
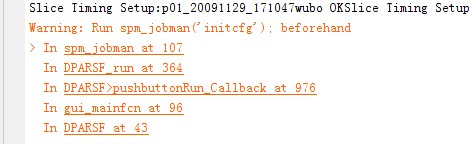
??? Error using ==> spm_slice_vol at 32
spm_slice_vol.c not compiled - see Makefile
Error in ==> spm_slice_timing at 213
slices(:,:,m) = spm_slice_vol(Vin(m),B,Vin(1).dim(1:2),1);
Error in ==> spm_config_slice_timing>slicetiming at 170
spm_slice_timing(P,Seq,refslice,timing)
Error in ==> spm_jobman>run_struct1 at 1474
feval(prog,val);
- Read more about troubles while processing DPARSF basic edition, never happened before
- 1 comment
- Log in or register to post comments
- 4863 reads