|
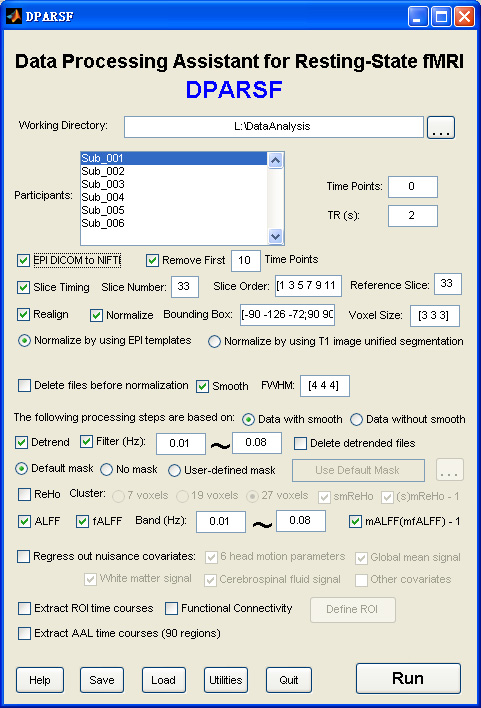
Data Processing Assistant for Resting-State fMRI (DPARSF) is a convenient plug-in software based on SPM and REST. You just need to arrange your DICOM files, and click a few buttons to set parameters, DPARSF will then give all the preprocessed (slice timing, realign, nomalize, smooth) data, FC, ReHo, ALFF and fALFF results. DPARSF can also create a report for excluding subjects with excessive head motion and generate a set of pictures for easily checking the effect of normalization. You can use DPARSF to extract AAL or ROI time courses (or extract Gray Matter Volume of AAL regions, command line only) efficiently if you want to perform small-world analysis. This software is very easy to use, just click on buttons if you are not sure what it means, popup tips would tell you what you need to do. You also can download a MULTIMEDIA COURSE to know more about how to use this software. Add DPARSF's directory to MATLAB's path and enter "DPARSF" in the command window of MATLAB to enjoy it.
The latest release is DPARSF_V1.0_100510.
DOWNLOAD
Multimedia Course: Data Processing of Resting-State fMRI |

