EPI T1 Dartel配准
老师好,我有个配准的问题 。 我做了T1和Dartel的配准,配准的效果都不好。 但是EPI的比较准! 那样我可以用EPI配准吗?在线等。
- Read more about EPI T1 Dartel配准
- 1 comment
- Log in or register to post comments
- 5681 reads
老师好,我有个配准的问题 。 我做了T1和Dartel的配准,配准的效果都不好。 但是EPI的比较准! 那样我可以用EPI配准吗?在线等。
老师您好,我们做配准的时候发现有些功能像的一些部分超出了脑外,不知道是否应该剔除,图像配准给出的图见附件。
非常感谢!
各位老师好:
我是一名神经外科研究生,想做一些与意识障碍有关的DTI联合rs-fMRI的研究,但我所在的医院只有1.5T GE磁共振,观看近几年的文章,基本都是3.0T的机器。2008年左右发表的文章有很多1.5T的,但不知道那些参数目前还能用否?
我做了几例预实验,采集静息态数据参数如下:
TR/TE=2000/40ms
Thickness/gap=5/1mm
Slices=24
Total time=372s(186 samples)
DTI数据参数如下:
TR/TE=9000/79.7ms
Thickness/gap=5/0mm
Slices=24
菜鸟求教:
静息态数据处理的时候,用voxel-wise全脑分析的时候,显著的CLUSTER包括本身这个种子点ROI怎么办?
谢谢各位老师!
老师好,我在一个教程上看到,在找到AC后,要在xyz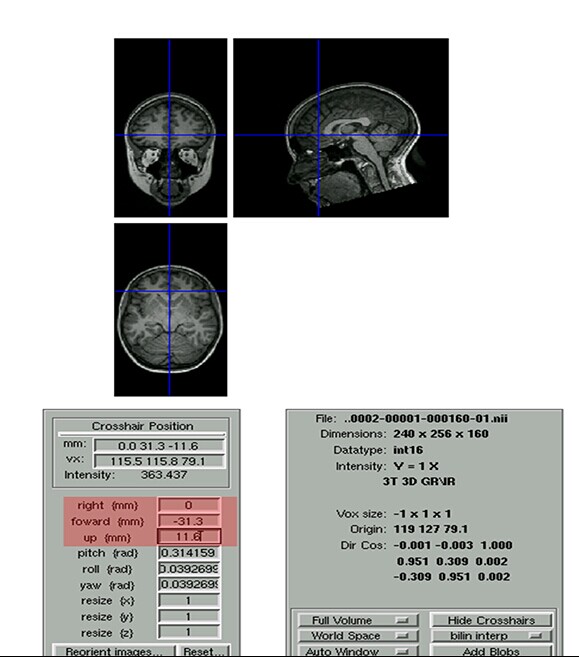 个地方,填的位置填上结果的相反数,这个在dparsfa里需要写吗? 看视频的意思是保持原来的 0 0 0 就好了?
个地方,填的位置填上结果的相反数,这个在dparsfa里需要写吗? 看视频的意思是保持原来的 0 0 0 就好了?
严老师:
您好!我在做完FC后,单样本T检验后能得到一组被试的整体的T-values,如果我想看每个个体某一脑区与ROI1的连接强度,是不是只要从zROI1FCMap中一个个体一个个体的提取相应脑区的值就可以了?这个值是应该用T-values表示还是应该用Z-values表示?
非常感谢!
初涉ICA有两点疑惑:
1. 臧老师提到说ICA的成分的意义是反应某一脑区同步性的高低。那么是否可以理解为其实质与ReHo的意义类似,只是不同的算法?
2. 在很多文献里面报告ICA的计算结果时,通常的表述是increased/decreased resting functional connectivity in ***,FC不应该指的是两者之间的一种关系吗?
求各位老师指点~!
多次在组会上被老板问道这个问题。所以想请教一下:
ReHo、ALFF的生理意义是什么?(物理意义是肯德尔一致性系数、BOLD信号某频段的能量大小)
看过臧老师最初发表的ReHo和ALFF的文章:ReHo与BOLD信号似乎都可以反应与手指运动相关的神经活动,ALFF可以区分睁眼闭眼的不同状态等。
不知道除了这两篇文章以外,最近几年是否有直接或者间接的研究证据阐释ReHo、ALFF与神经电活动之间的关系?
比方说:有没有ReHo值/ALFF值的降低或升高反映了神经活动水平的上升?之类的证据?
真的是被老板问了好多次了。所以请教一下各位老师对静息态指标(ICA、ReHo、ALFF、FC、小世界网络等)究竟能告诉我们什么?
弱弱地问一下: 为什么大家一般会用smReHo作为统计的指标?而不是原始的ReHo值?
开组会讨论数据结果的时候,老板提的问题:
1. smReHo是相对值,而ReHo是原始的绝对值,统计的时候为什么用相对值,而不用绝对值呢?
2. 如果全脑平均的ReHo值在不同组的被试之间差异显著(即全脑均值本身存在组间差异),那还可以使用smReHo做统计么?
3. 比较smReHo(相对值)的组间差异的意义是什么呢?
谢谢各位老师!^^