gift成分导出
请问张老师,在GIFT里面如何将成分导出来呢?
- Read more about gift成分导出
- 7 comments
- Log in or register to post comments
- 12777 reads
请问张老师,在GIFT里面如何将成分导出来呢?
Dear experts,
I'm quite new with the toolkit REST,
and I have a doubt:
I have two groups (paitents-healthy controls) and I have 2 resting-state runs for each subject.
Once I performed ReHo (and ALFF/fALFF) calculation for each single run,
各位老师好:
我使用静息态的数据做功能连接,利用Voxel-based的方法研究M1区的功能连接,种子点选择左右两侧的M1区,出来的全脑功能连接图取了阈值之后总是会出现两侧的枕叶区域,我们的数据都是闭眼扫静息态的,我查的做运动功能连接的文献里面基本上都没提到枕叶区域,不知道是他们的结果就没枕叶还是他们没报道。在我矫正很严格的情况下枕叶还是存在,不知该如何解释这种情况?是不是比如我只关心和运动相关的脑区,但是出来一些明显和运动无关的脑区可以不用报出来或解释,请指导~~
大家好!我想请教各位老师,怎样才能分出扫描顺序呢?
大家好!我是一名刚开始接触resting-state fMRI 的学生,我这里有一些核磁数据,但不知道怎么分出是自上而下扫的还是相反方向,也分不出是隔层扫的还是顺序扫的,我想请教大家有没有什么软件之类的可以自动分出。
Error using flipud (line 19)
X must be a 2-D matrix.
Error in rest_ImgCal_gui>img_cal (line 481)
eval(['Y = ' f ';']);
Error in rest_ImgCal_gui>btnCompute_Callback (line 228)
img_cal(OutDir,OutPut,Function,Gmag,data);
Error in gui_mainfcn (line 96)
各位老师好:
请问DPARSFA最新版本计算degree cenrality得到的是global FCD还是local FCD?
另外,network centrality与node centrality 或nodal centrality是否为同义词?
感谢您的热心解答。
Xiao-dong Zhang
jinling hospital
2014-4-3
张老师你好:
我有两个问题想咨询下
Dear rsFC analysis experts,
I have some questions with respect to DC analysis.
各位老师,各位朋友: 我在处理rest fmri时,发现有好几个数据本来原始T1像是有小脑的,但coreg to Fun后,小脑就不全了,请问是怎么回事呢?该怎么处理呢?
co-T1正常情况:
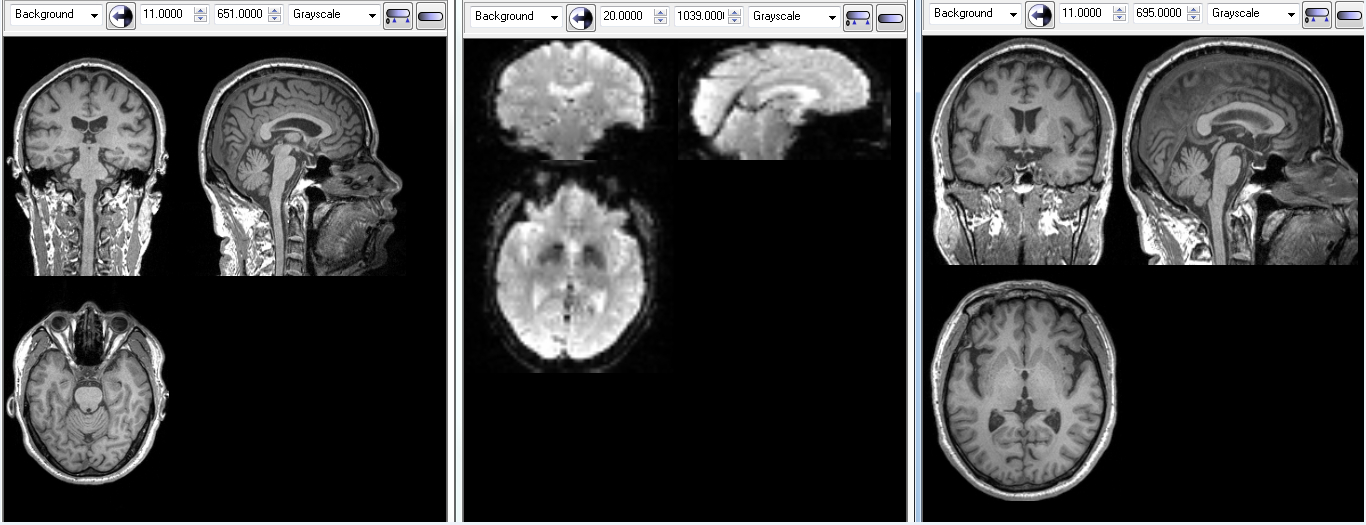
co-T1小脑不全情况:
.jpg)