dparsf并行报错
老师:
matlab并行处理的时候总是报这样的错误?请问是什么原因。在做完头动矫正之后。
- Read more about dparsf并行报错
- 2 comments
- Log in or register to post comments
- 7750 reads
老师:
matlab并行处理的时候总是报这样的错误?请问是什么原因。在做完头动矫正之后。
大家好:
我在处理同一批(52个被试)静息态数据时,用EPI模板进行空间配准化,空间标准化之后的图像没有问题。而用DARTEL模板进行空间标准化,标准化之后的数据,基本每个被试都发现有点信号丢失,请分析一下是预处理不对的原因还是什么?并附预处理的过程及分别对应同一个被试,同一个时间点的图像。假如我用EPI模板去空间标准化,在后面的统计计算时用DARTEL的灰质数据制做灰质的MASK会不会有问题?非常感谢!
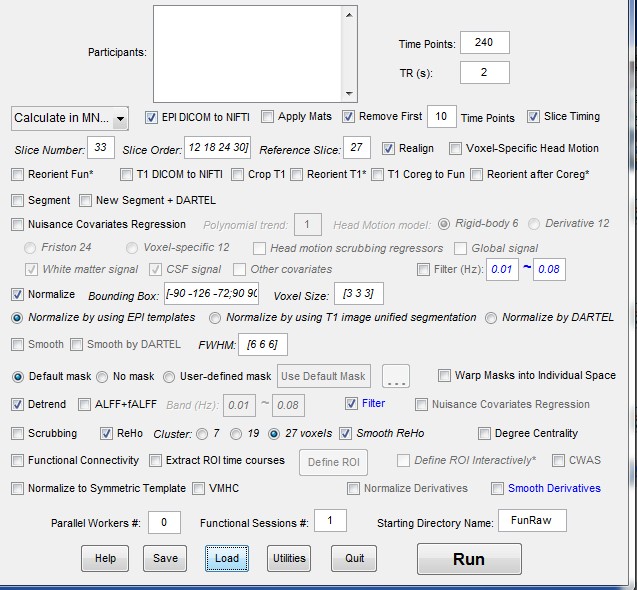
Hi,
After running the preprocessing steps within DPARSF, I got the following error:
??? Error using ==> set
请问一下,在rest统计模块中单样本t检验中,假设以海马为ROI的FC来说,结果显示有正有负,分别代表什么含义?
取0.05的阈值之后,是不是留下来的都是有统计意义的?
colorbar选12,那红色和蓝色代表什么,正性连接,负性连接?还是两者没有区别?
各位老师好:
我目前在进行一个动物的数据分析,需要将动物模板替换VBM8自带的模板。但VBM8自带的TPM模板,这是一个4d的数据,包括6个三维数据。
我的模板分为T1,T2,CSF,GM,WM,将TPM换成TI,或者灰质的模板,都无法运行。提示:can't get volum information for"我输入的模板地址"。请问我该如何添加动物模板。
谢谢赐教!
各位老师好!
启动rest时报错如下(在matalab2014a上运行):
警告: 未找到文件或者权限被拒绝
> In rest at 51
错误使用 diary
C:\Program Files\MATLAB\fMRITool\REST_V1.8_130615\rest.log: 无法打开文件: 权限被拒绝。
出错 rest (line 53)
diary(fullfile(rest_misc('WhereIsREST'), 'rest.log'));
反复看了rest第51行,感觉没什么错误,不知道为什么?
谢谢
报错内容
Warning: Matrix is singular to working precision.
> In DynamicBC_fls_FC>wgr_fls at 92
In DynamicBC_fls_FC>(parfor body) at 21
In parallel_function at 475
In DynamicBC_fls_FC at 17
In DynamicBC_run at 267
In DynamicBC>wgr_run_check at 1119
设置情况如下
win2008, Matlab 2011b
出错文件如下
Error using DynamicBC_sliding_window_FC>wgr_correl_FCD (line 220)
Error: The expression to the left of the equals sign is not a valid target for an assignment.
Error in DynamicBC_sliding_window_FC (line 41)
出错内容如下 win2008,Matlab2011b ,DPARSF V2.2_121225,Rest 1.8
任务设置文件见附件
多谢指教!
Error using y_ExtractROISignal (line 188)
Wrong ROI definition, please check:
D:\FTP\Process_new\Masks\20121121002_BrainMask_05_91x109x91.nii
Error in DPARSFA_run>(parfor body) (line 1851)
大家好,